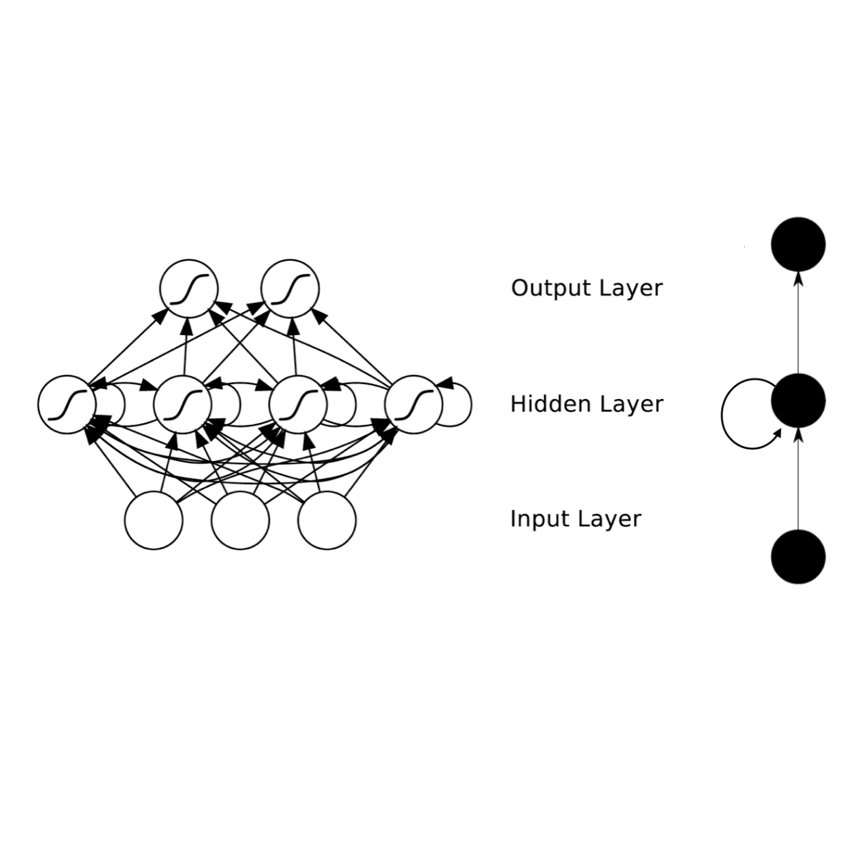
Languages have long been described according to their perceived rhythmic attributes. The associated typologies are of interest in psycholinguistics as they partly predict newborns' abilities to discriminate between languages and provide insights into how adult listeners process non-native languages. Despite the relative success of rhythm metrics in supporting the existence of linguistic rhythmic classes, quantitative studies have yet to capture the full complexity of temporal regularities associated with speech rhythm. We argue that deep learning offers a powerful pattern-recognition approach to advance the characterization of the acoustic bases of speech rhythm. To explore this hypothesis, we trained a medium-sized recurrent neural network on a language identification task over a large database of speech recordings in 21 languages. The network had access to the amplitude envelopes and a variable identifying the voiced segments, assuming that this signal would poorly convey phonetic information but preserve prosodic features. The network was able to identify the language of 10-second recordings in 40% of the cases, and the language was in the top-3 guesses in two-thirds of the cases. Visualization methods show that representations built from the network activations are consistent with speech rhythm typologies, although the resulting maps are more complex than two separated clusters between stress and syllable-timed languages. We further analyzed the model by identifying correlations between network activations and known speech rhythm metrics. The findings illustrate the potential of deep learning tools to advance our understanding of speech rhythm through the identification and exploration of linguistically relevant acoustic feature spaces.
相關內容
Efficient and accurate algorithms are necessary to reconstruct particles in the highly granular detectors anticipated at the High-Luminosity Large Hadron Collider and the Future Circular Collider. We study scalable machine learning models for event reconstruction in electron-positron collisions based on a full detector simulation. Particle-flow reconstruction can be formulated as a supervised learning task using tracks and calorimeter clusters. We compare a graph neural network and kernel-based transformer and demonstrate that we can avoid quadratic operations while achieving realistic reconstruction. We show that hyperparameter tuning significantly improves the performance of the models. The best graph neural network model shows improvement in the jet transverse momentum resolution by up to 50% compared to the rule-based algorithm. The resulting model is portable across Nvidia, AMD and Habana hardware. Accurate and fast machine-learning based reconstruction can significantly improve future measurements at colliders.
This paper presents a novel stochastic optimisation methodology to perform empirical Bayesian inference in semi-blind image deconvolution problems. Given a blurred image and a parametric class of possible operators, the proposed optimisation approach automatically calibrates the parameters of the blur model by maximum marginal likelihood estimation, followed by (non-blind) image deconvolution by maximum-a-posteriori estimation conditionally to the estimated model parameters. In addition to the blur model, the proposed approach also automatically calibrates the noise variance as well as any regularisation parameters. The marginal likelihood of the blur, noise variance, and regularisation parameters is generally computationally intractable, as it requires calculating several integrals over the entire solution space. Our approach addresses this difficulty by using a stochastic approximation proximal gradient optimisation scheme, which iteratively solves such integrals by using a Moreau-Yosida regularised unadjusted Langevin Markov chain Monte Carlo algorithm. This optimisation strategy can be easily and efficiently applied to any model that is log-concave, and by using the same gradient and proximal operators that are required to compute the maximum-a-posteriori solution by convex optimisation. We provide convergence guarantees for the proposed optimisation scheme under realistic and easily verifiable conditions and subsequently demonstrate the effectiveness of the approach with a series of deconvolution experiments and comparisons with alternative strategies from the state of the art.
We present a new methodology for decomposing flows with multiple transports that further extends the shifted proper orthogonal decomposition (sPOD). The sPOD tries to approximate transport-dominated flows by a sum of co-moving data fields. The proposed methods stem from sPOD but optimize the co-moving fields directly and penalize their nuclear norm to promote low rank of the individual data in the decomposition. Furthermore, we add a robustness term to the decomposition that can deal with interpolation error and data noises. Leveraging tools from convex optimization, we derive three proximal algorithms to solve the decomposition problem. We report a numerical comparison with existing methods against synthetic data benchmarks and then show the separation ability of our methods on 1D and 2D incompressible and reactive flows. The resulting methodology is the basis of a new analysis paradigm that results in the same interpretability as the POD for the individual co-moving fields.
Protein design requires a deep understanding of the inherent complexities of the protein universe. While many efforts lean towards conditional generation or focus on specific families of proteins, the foundational task of unconditional generation remains underexplored and undervalued. Here, we explore this pivotal domain, introducing DiMA, a model that leverages continuous diffusion on embeddings derived from the protein language model, ESM-2, to generate amino acid sequences. DiMA surpasses leading solutions, including autoregressive transformer-based and discrete diffusion models, and we quantitatively illustrate the impact of the design choices that lead to its superior performance. We extensively evaluate the quality, diversity, distribution similarity, and biological relevance of the generated sequences using multiple metrics across various modalities. Our approach consistently produces novel, diverse protein sequences that accurately reflect the inherent structural and functional diversity of the protein space. This work advances the field of protein design and sets the stage for conditional models by providing a robust framework for scalable and high-quality protein sequence generation.
We show that the known list-decoding algorithms for univariate multiplicity and folded Reed-Solomon (FRS) codes can be made to run in nearly-linear time. This yields, to our knowledge, the first known family of codes that can be decoded in nearly linear time, even as they approach the list decoding capacity. Univariate multiplicity codes and FRS codes are natural variants of Reed-Solomon codes that were discovered and studied for their applications to list-decoding. It is known that for every $\epsilon >0$, and rate $R \in (0,1)$, there exist explicit families of these codes that have rate $R$ and can be list-decoded from a $(1-R-\epsilon)$ fraction of errors with constant list size in polynomial time (Guruswami & Wang (IEEE Trans. Inform. Theory, 2013) and Kopparty, Ron-Zewi, Saraf & Wootters (SIAM J. Comput. 2023)). In this work, we present randomized algorithms that perform the above tasks in nearly linear time. Our algorithms have two main components. The first builds upon the lattice-based approach of Alekhnovich (IEEE Trans. Inf. Theory 2005), who designed a nearly linear time list-decoding algorithm for Reed-Solomon codes approaching the Johnson radius. As part of the second component, we design nearly-linear time algorithms for two natural algebraic problems. The first algorithm solves linear differential equations of the form $Q\left(x, f(x), \frac{df}{dx}, \dots,\frac{d^m f}{dx^m}\right) \equiv 0$ where $Q$ has the form $Q(x,y_0,\dots,y_m) = \tilde{Q}(x) + \sum_{i = 0}^m Q_i(x)\cdot y_i$. The second solves functional equations of the form $Q\left(x, f(x), f(\gamma x), \dots,f(\gamma^m x)\right) \equiv 0$ where $\gamma$ is a high-order field element. These algorithms can be viewed as generalizations of classical algorithms of Sieveking (Computing 1972) and Kung (Numer. Math. 1974) for computing the modular inverse of a power series, and might be of independent interest.
Necessary and sufficient conditions of uniform consistency are explored. A hypothesis is simple. Nonparametric sets of alternatives are bounded convex sets in $\mathbb{L}_p$, $p >1$ with "small" balls deleted. The "small" balls have the center at the point of hypothesis and radii of balls tend to zero as sample size increases. For problem of hypothesis testing on a density, we show that, for the sets of alternatives, there are uniformly consistent tests for some sequence of radii of the balls, if and only if, convex set is relatively compact. The results are established for problem of hypothesis testing on a density, for signal detection in Gaussian white noise, for linear ill-posed problems with random Gaussian noise and so on.
Deployable polyhedrons can transform between Platonic and Archimedean polyhedrons to meet the demands of various engineering applications. However, the existing design solutions are often with multiple degrees of freedom and complicated mechanism links and joints, which greatly limited their potential in practice. Combining the fundamentals of solid geometry and mechanism kinematics, this paper proposes a family of kirigami Archimedean polyhedrons based on the N-fold-symmetric loops of spatial 7R linkage, which perform one-DOF radial transformation following tetrahedral, octahedral, or icosahedral symmetry. Moreover, in each symmetric polyhedral group, three different transforming paths can be achieved from one identical deployed configuration. We also demonstrated that such design strategy can be readily applied to polyhedral tessellation. This work provides a family of rich solutions for deployable polyhedrons to facilitate their applications in aerospace exploration, architecture, metamaterials and so on.
We hypothesize that due to the greedy nature of learning in multi-modal deep neural networks, these models tend to rely on just one modality while under-fitting the other modalities. Such behavior is counter-intuitive and hurts the models' generalization, as we observe empirically. To estimate the model's dependence on each modality, we compute the gain on the accuracy when the model has access to it in addition to another modality. We refer to this gain as the conditional utilization rate. In the experiments, we consistently observe an imbalance in conditional utilization rates between modalities, across multiple tasks and architectures. Since conditional utilization rate cannot be computed efficiently during training, we introduce a proxy for it based on the pace at which the model learns from each modality, which we refer to as the conditional learning speed. We propose an algorithm to balance the conditional learning speeds between modalities during training and demonstrate that it indeed addresses the issue of greedy learning. The proposed algorithm improves the model's generalization on three datasets: Colored MNIST, Princeton ModelNet40, and NVIDIA Dynamic Hand Gesture.
Graph representation learning for hypergraphs can be used to extract patterns among higher-order interactions that are critically important in many real world problems. Current approaches designed for hypergraphs, however, are unable to handle different types of hypergraphs and are typically not generic for various learning tasks. Indeed, models that can predict variable-sized heterogeneous hyperedges have not been available. Here we develop a new self-attention based graph neural network called Hyper-SAGNN applicable to homogeneous and heterogeneous hypergraphs with variable hyperedge sizes. We perform extensive evaluations on multiple datasets, including four benchmark network datasets and two single-cell Hi-C datasets in genomics. We demonstrate that Hyper-SAGNN significantly outperforms the state-of-the-art methods on traditional tasks while also achieving great performance on a new task called outsider identification. Hyper-SAGNN will be useful for graph representation learning to uncover complex higher-order interactions in different applications.

Recent advances in 3D fully convolutional networks (FCN) have made it feasible to produce dense voxel-wise predictions of volumetric images. In this work, we show that a multi-class 3D FCN trained on manually labeled CT scans of several anatomical structures (ranging from the large organs to thin vessels) can achieve competitive segmentation results, while avoiding the need for handcrafting features or training class-specific models. To this end, we propose a two-stage, coarse-to-fine approach that will first use a 3D FCN to roughly define a candidate region, which will then be used as input to a second 3D FCN. This reduces the number of voxels the second FCN has to classify to ~10% and allows it to focus on more detailed segmentation of the organs and vessels. We utilize training and validation sets consisting of 331 clinical CT images and test our models on a completely unseen data collection acquired at a different hospital that includes 150 CT scans, targeting three anatomical organs (liver, spleen, and pancreas). In challenging organs such as the pancreas, our cascaded approach improves the mean Dice score from 68.5 to 82.2%, achieving the highest reported average score on this dataset. We compare with a 2D FCN method on a separate dataset of 240 CT scans with 18 classes and achieve a significantly higher performance in small organs and vessels. Furthermore, we explore fine-tuning our models to different datasets. Our experiments illustrate the promise and robustness of current 3D FCN based semantic segmentation of medical images, achieving state-of-the-art results. Our code and trained models are available for download: //github.com/holgerroth/3Dunet_abdomen_cascade.