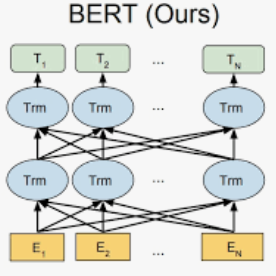
We present an automatic COVID1-19 diagnosis framework from lung CT-scan slice images. In this framework, the slice images of a CT-scan volume are first proprocessed using segmentation techniques to filter out images of closed lung, and to remove the useless background. Then a resampling method is used to select one or multiple sets of a fixed number of slice images for training and validation. A 3D CNN network with BERT is used to classify this set of selected slice images. In this network, an embedding feature is also extracted. In cases where there are more than one set of slice images in a volume, the features of all sets are extracted and pooled into a global feature vector for the whole CT-scan volume. A simple multiple-layer perceptron (MLP) network is used to further classify the aggregated feature vector. The models are trained and evaluated on the provided training and validation datasets. On the validation dataset, the accuracy is 0.9278 and the F1 score is 0.9261.
相關內容
The COVID-19 pandemic continues to have a devastating effect on the health and well-being of the global population. A critical step in the fight against COVID-19 is effective screening of infected patients, with one of the key screening approaches being radiological imaging using chest radiography. Motivated by this, a number of artificial intelligence (AI) systems based on deep learning have been proposed and results have been shown to be quite promising in terms of accuracy in detecting patients infected with COVID-19 using chest radiography images. However, to the best of the authors' knowledge, these developed AI systems have been closed source and unavailable to the research community for deeper understanding and extension, and unavailable for public access and use. Therefore, in this study we introduce COVID-Net, a deep convolutional neural network design tailored for the detection of COVID-19 cases from chest radiography images that is open source and available to the general public. We also describe the chest radiography dataset leveraged to train COVID-Net, which we will refer to as COVIDx and is comprised of 5941 posteroanterior chest radiography images across 2839 patient cases from two open access data repositories. Furthermore, we investigate how COVID-Net makes predictions using an explainability method in an attempt to gain deeper insights into critical factors associated with COVID cases, which can aid clinicians in improved screening. By no means a production-ready solution, the hope is that the open access COVID-Net, along with the description on constructing the open source COVIDx dataset, will be leveraged and build upon by both researchers and citizen data scientists alike to accelerate the development of highly accurate yet practical deep learning solutions for detecting COVID-19 cases and accelerate treatment of those who need it the most.
We study the impact of neural networks in text classification. Our focus is on training deep neural networks with proper weight initialization and greedy layer-wise pretraining. Results are compared with 1-layer neural networks and Support Vector Machines. We work with a dataset of labeled messages from the Twitter microblogging service and aim to predict weather conditions. A feature extraction procedure specific for the task is proposed, which applies dimensionality reduction using Latent Semantic Analysis. Our results show that neural networks outperform Support Vector Machines with Gaussian kernels, noticing performance gains from introducing additional hidden layers with nonlinearities. The impact of using Nesterov's Accelerated Gradient in backpropagation is also studied. We conclude that deep neural networks are a reasonable approach for text classification and propose further ideas to improve performance.
Detecting objects in aerial images is challenging for at least two reasons: (1) target objects like pedestrians are very small in pixels, making them hardly distinguished from surrounding background; and (2) targets are in general sparsely and non-uniformly distributed, making the detection very inefficient. In this paper, we address both issues inspired by observing that these targets are often clustered. In particular, we propose a Clustered Detection (ClusDet) network that unifies object clustering and detection in an end-to-end framework. The key components in ClusDet include a cluster proposal sub-network (CPNet), a scale estimation sub-network (ScaleNet), and a dedicated detection network (DetecNet). Given an input image, CPNet produces object cluster regions and ScaleNet estimates object scales for these regions. Then, each scale-normalized cluster region is fed into DetecNet for object detection. ClusDet has several advantages over previous solutions: (1) it greatly reduces the number of chips for final object detection and hence achieves high running time efficiency, (2) the cluster-based scale estimation is more accurate than previously used single-object based ones, hence effectively improves the detection for small objects, and (3) the final DetecNet is dedicated for clustered regions and implicitly models the prior context information so as to boost detection accuracy. The proposed method is tested on three popular aerial image datasets including VisDrone, UAVDT and DOTA. In all experiments, ClusDet achieves promising performance in comparison with state-of-the-art detectors. Code will be available in \url{//github.com/fyangneil}.
Deep neural network models used for medical image segmentation are large because they are trained with high-resolution three-dimensional (3D) images. Graphics processing units (GPUs) are widely used to accelerate the trainings. However, the memory on a GPU is not large enough to train the models. A popular approach to tackling this problem is patch-based method, which divides a large image into small patches and trains the models with these small patches. However, this method would degrade the segmentation quality if a target object spans multiple patches. In this paper, we propose a novel approach for 3D medical image segmentation that utilizes the data-swapping, which swaps out intermediate data from GPU memory to CPU memory to enlarge the effective GPU memory size, for training high-resolution 3D medical images without patching. We carefully tuned parameters in the data-swapping method to obtain the best training performance for 3D U-Net, a widely used deep neural network model for medical image segmentation. We applied our tuning to train 3D U-Net with full-size images of 192 x 192 x 192 voxels in brain tumor dataset. As a result, communication overhead, which is the most important issue, was reduced by 17.1%. Compared with the patch-based method for patches of 128 x 128 x 128 voxels, our training for full-size images achieved improvement on the mean Dice score by 4.48% and 5.32 % for detecting whole tumor sub-region and tumor core sub-region, respectively. The total training time was reduced from 164 hours to 47 hours, resulting in 3.53 times of acceleration.
Radiologist is "doctor's doctor", biomedical image segmentation plays a central role in quantitative analysis, clinical diagnosis, and medical intervention. In the light of the fully convolutional networks (FCN) and U-Net, deep convolutional networks (DNNs) have made significant contributions in biomedical image segmentation applications. In this paper, based on U-Net, we propose MDUnet, a multi-scale densely connected U-net for biomedical image segmentation. we propose three different multi-scale dense connections for U shaped architectures encoder, decoder and across them. The highlights of our architecture is directly fuses the neighboring different scale feature maps from both higher layers and lower layers to strengthen feature propagation in current layer. Which can largely improves the information flow encoder, decoder and across them. Multi-scale dense connections, which means containing shorter connections between layers close to the input and output, also makes much deeper U-net possible. We adopt the optimal model based on the experiment and propose a novel Multi-scale Dense U-Net (MDU-Net) architecture with quantization. Which reduce overfitting in MDU-Net for better accuracy. We evaluate our purpose model on the MICCAI 2015 Gland Segmentation dataset (GlaS). The three multi-scale dense connections improve U-net performance by up to 1.8% on test A and 3.5% on test B in the MICCAI Gland dataset. Meanwhile the MDU-net with quantization achieves the superiority over U-Net performance by up to 3% on test A and 4.1% on test B.
Modern CNN-based object detectors rely on bounding box regression and non-maximum suppression to localize objects. While the probabilities for class labels naturally reflect classification confidence, localization confidence is absent. This makes properly localized bounding boxes degenerate during iterative regression or even suppressed during NMS. In the paper we propose IoU-Net learning to predict the IoU between each detected bounding box and the matched ground-truth. The network acquires this confidence of localization, which improves the NMS procedure by preserving accurately localized bounding boxes. Furthermore, an optimization-based bounding box refinement method is proposed, where the predicted IoU is formulated as the objective. Extensive experiments on the MS-COCO dataset show the effectiveness of IoU-Net, as well as its compatibility with and adaptivity to several state-of-the-art object detectors.
Deep neural network architectures have traditionally been designed and explored with human expertise in a long-lasting trial-and-error process. This process requires huge amount of time, expertise, and resources. To address this tedious problem, we propose a novel algorithm to optimally find hyperparameters of a deep network architecture automatically. We specifically focus on designing neural architectures for medical image segmentation task. Our proposed method is based on a policy gradient reinforcement learning for which the reward function is assigned a segmentation evaluation utility (i.e., dice index). We show the efficacy of the proposed method with its low computational cost in comparison with the state-of-the-art medical image segmentation networks. We also present a new architecture design, a densely connected encoder-decoder CNN, as a strong baseline architecture to apply the proposed hyperparameter search algorithm. We apply the proposed algorithm to each layer of the baseline architectures. As an application, we train the proposed system on cine cardiac MR images from Automated Cardiac Diagnosis Challenge (ACDC) MICCAI 2017. Starting from a baseline segmentation architecture, the resulting network architecture obtains the state-of-the-art results in accuracy without performing any trial-and-error based architecture design approaches or close supervision of the hyperparameters changes.

Recent advances in 3D fully convolutional networks (FCN) have made it feasible to produce dense voxel-wise predictions of volumetric images. In this work, we show that a multi-class 3D FCN trained on manually labeled CT scans of several anatomical structures (ranging from the large organs to thin vessels) can achieve competitive segmentation results, while avoiding the need for handcrafting features or training class-specific models. To this end, we propose a two-stage, coarse-to-fine approach that will first use a 3D FCN to roughly define a candidate region, which will then be used as input to a second 3D FCN. This reduces the number of voxels the second FCN has to classify to ~10% and allows it to focus on more detailed segmentation of the organs and vessels. We utilize training and validation sets consisting of 331 clinical CT images and test our models on a completely unseen data collection acquired at a different hospital that includes 150 CT scans, targeting three anatomical organs (liver, spleen, and pancreas). In challenging organs such as the pancreas, our cascaded approach improves the mean Dice score from 68.5 to 82.2%, achieving the highest reported average score on this dataset. We compare with a 2D FCN method on a separate dataset of 240 CT scans with 18 classes and achieve a significantly higher performance in small organs and vessels. Furthermore, we explore fine-tuning our models to different datasets. Our experiments illustrate the promise and robustness of current 3D FCN based semantic segmentation of medical images, achieving state-of-the-art results. Our code and trained models are available for download: //github.com/holgerroth/3Dunet_abdomen_cascade.
This paper reports Deep LOGISMOS approach to 3D tumor segmentation by incorporating boundary information derived from deep contextual learning to LOGISMOS - layered optimal graph image segmentation of multiple objects and surfaces. Accurate and reliable tumor segmentation is essential to tumor growth analysis and treatment selection. A fully convolutional network (FCN), UNet, is first trained using three adjacent 2D patches centered at the tumor, providing contextual UNet segmentation and probability map for each 2D patch. The UNet segmentation is then refined by Gaussian Mixture Model (GMM) and morphological operations. The refined UNet segmentation is used to provide the initial shape boundary to build a segmentation graph. The cost for each node of the graph is determined by the UNet probability maps. Finally, a max-flow algorithm is employed to find the globally optimal solution thus obtaining the final segmentation. For evaluation, we applied the method to pancreatic tumor segmentation on a dataset of 51 CT scans, among which 30 scans were used for training and 21 for testing. With Deep LOGISMOS, DICE Similarity Coefficient (DSC) and Relative Volume Difference (RVD) reached 83.2+-7.8% and 18.6+-17.4% respectively, both are significantly improved (p<0.05) compared with contextual UNet and/or LOGISMOS alone.
Recent advance in fluorescence microscopy enables acquisition of 3D image volumes with better quality and deeper penetration into tissue. Segmentation is a required step to characterize and analyze biological structures in the images. 3D segmentation using deep learning has achieved promising results in microscopy images. One issue is that deep learning techniques require a large set of groundtruth data which is impractical to annotate manually for microscopy volumes. This paper describes a 3D nuclei segmentation method using 3D convolutional neural networks. A set of synthetic volumes and the corresponding groundtruth volumes are generated automatically using a generative adversarial network. Segmentation results demonstrate that our proposed method is capable of segmenting nuclei successfully in 3D for various data sets.