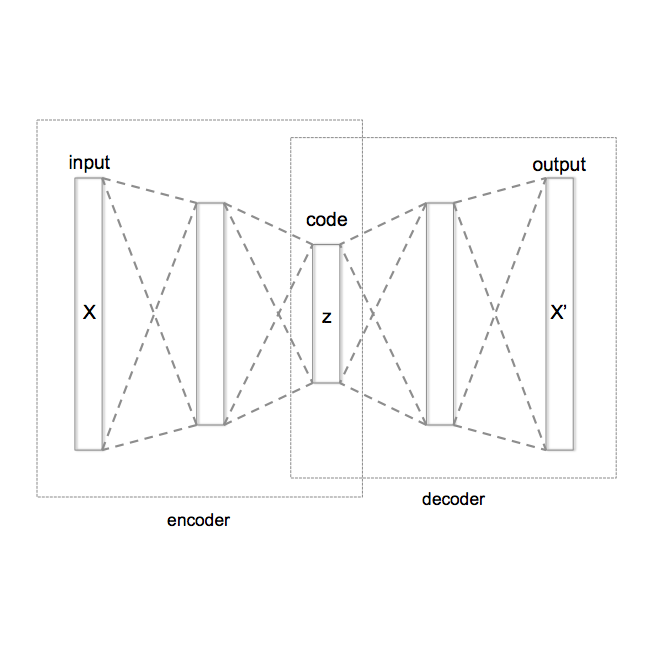
Graph structured data are abundant in the real world. Among different graph types, directed acyclic graphs (DAGs) are of particular interest to machine learning researchers, as many machine learning models are realized as computations on DAGs, including neural networks and Bayesian networks. In this paper, we study deep generative models for DAGs, and propose a novel DAG variational autoencoder (D-VAE). To encode DAGs into the latent space, we leverage graph neural networks. We propose an asynchronous message passing scheme that allows encoding the computations on DAGs, rather than using existing simultaneous message passing schemes to encode local graph structures. We demonstrate the effectiveness of our proposed D-VAE through two tasks: neural architecture search and Bayesian network structure learning. Experiments show that our model not only generates novel and valid DAGs, but also produces a smooth latent space that facilitates searching for DAGs with better performance through Bayesian optimization.
相關內容
Modeling multivariate time series has long been a subject that has attracted researchers from a diverse range of fields including economics, finance, and traffic. A basic assumption behind multivariate time series forecasting is that its variables depend on one another but, upon looking closely, it is fair to say that existing methods fail to fully exploit latent spatial dependencies between pairs of variables. In recent years, meanwhile, graph neural networks (GNNs) have shown high capability in handling relational dependencies. GNNs require well-defined graph structures for information propagation which means they cannot be applied directly for multivariate time series where the dependencies are not known in advance. In this paper, we propose a general graph neural network framework designed specifically for multivariate time series data. Our approach automatically extracts the uni-directed relations among variables through a graph learning module, into which external knowledge like variable attributes can be easily integrated. A novel mix-hop propagation layer and a dilated inception layer are further proposed to capture the spatial and temporal dependencies within the time series. The graph learning, graph convolution, and temporal convolution modules are jointly learned in an end-to-end framework. Experimental results show that our proposed model outperforms the state-of-the-art baseline methods on 3 of 4 benchmark datasets and achieves on-par performance with other approaches on two traffic datasets which provide extra structural information.
Representation learning on a knowledge graph (KG) is to embed entities and relations of a KG into low-dimensional continuous vector spaces. Early KG embedding methods only pay attention to structured information encoded in triples, which would cause limited performance due to the structure sparseness of KGs. Some recent attempts consider paths information to expand the structure of KGs but lack explainability in the process of obtaining the path representations. In this paper, we propose a novel Rule and Path-based Joint Embedding (RPJE) scheme, which takes full advantage of the explainability and accuracy of logic rules, the generalization of KG embedding as well as the supplementary semantic structure of paths. Specifically, logic rules of different lengths (the number of relations in rule body) in the form of Horn clauses are first mined from the KG and elaborately encoded for representation learning. Then, the rules of length 2 are applied to compose paths accurately while the rules of length 1 are explicitly employed to create semantic associations among relations and constrain relation embeddings. Besides, the confidence level of each rule is also considered in optimization to guarantee the availability of applying the rule to representation learning. Extensive experimental results illustrate that RPJE outperforms other state-of-the-art baselines on KG completion task, which also demonstrate the superiority of utilizing logic rules as well as paths for improving the accuracy and explainability of representation learning.
Graph autoencoders (AE) and variational autoencoders (VAE) recently emerged as powerful node embedding methods. In particular, graph AE and VAE were successfully leveraged to tackle the challenging link prediction problem, aiming at figuring out whether some pairs of nodes from a graph are connected by unobserved edges. However, these models focus on undirected graphs and therefore ignore the potential direction of the link, which is limiting for numerous real-life applications. In this paper, we extend the graph AE and VAE frameworks to address link prediction in directed graphs. We present a new gravity-inspired decoder scheme that can effectively reconstruct directed graphs from a node embedding. We empirically evaluate our method on three different directed link prediction tasks, for which standard graph AE and VAE perform poorly. We achieve competitive results on three real-world graphs, outperforming several popular baselines.
We present new intuitions and theoretical assessments of the emergence of disentangled representation in variational autoencoders. Taking a rate-distortion theory perspective, we show the circumstances under which representations aligned with the underlying generative factors of variation of data emerge when optimising the modified ELBO bound in $\beta$-VAE, as training progresses. From these insights, we propose a modification to the training regime of $\beta$-VAE, that progressively increases the information capacity of the latent code during training. This modification facilitates the robust learning of disentangled representations in $\beta$-VAE, without the previous trade-off in reconstruction accuracy.
Inferring missing links in knowledge graphs (KG) has attracted a lot of attention from the research community. In this paper, we tackle a practical query answering task involving predicting the relation of a given entity pair. We frame this prediction problem as an inference problem in a probabilistic graphical model and aim at resolving it from a variational inference perspective. In order to model the relation between the query entity pair, we assume that there exists an underlying latent variable (paths connecting two nodes) in the KG, which carries the equivalent semantics of their relations. However, due to the intractability of connections in large KGs, we propose to use variation inference to maximize the evidence lower bound. More specifically, our framework (\textsc{Diva}) is composed of three modules, i.e. a posterior approximator, a prior (path finder), and a likelihood (path reasoner). By using variational inference, we are able to incorporate them closely into a unified architecture and jointly optimize them to perform KG reasoning. With active interactions among these sub-modules, \textsc{Diva} is better at handling noise and coping with more complex reasoning scenarios. In order to evaluate our method, we conduct the experiment of the link prediction task on multiple datasets and achieve state-of-the-art performances on both datasets.
We propose a flipped-Adversarial AutoEncoder (FAAE) that simultaneously trains a generative model G that maps an arbitrary latent code distribution to a data distribution and an encoder E that embodies an "inverse mapping" that encodes a data sample into a latent code vector. Unlike previous hybrid approaches that leverage adversarial training criterion in constructing autoencoders, FAAE minimizes re-encoding errors in the latent space and exploits adversarial criterion in the data space. Experimental evaluations demonstrate that the proposed framework produces sharper reconstructed images while at the same time enabling inference that captures rich semantic representation of data.

Deep metric learning has been demonstrated to be highly effective in learning semantic representation and encoding information that can be used to measure data similarity, by relying on the embedding learned from metric learning. At the same time, variational autoencoder (VAE) has widely been used to approximate inference and proved to have a good performance for directed probabilistic models. However, for traditional VAE, the data label or feature information are intractable. Similarly, traditional representation learning approaches fail to represent many salient aspects of the data. In this project, we propose a novel integrated framework to learn latent embedding in VAE by incorporating deep metric learning. The features are learned by optimizing a triplet loss on the mean vectors of VAE in conjunction with standard evidence lower bound (ELBO) of VAE. This approach, which we call Triplet based Variational Autoencoder (TVAE), allows us to capture more fine-grained information in the latent embedding. Our model is tested on MNIST data set and achieves a high triplet accuracy of 95.60% while the traditional VAE (Kingma & Welling, 2013) achieves triplet accuracy of 75.08%.
Methods that learn representations of nodes in a graph play a critical role in network analysis since they enable many downstream learning tasks. We propose Graph2Gauss - an approach that can efficiently learn versatile node embeddings on large scale (attributed) graphs that show strong performance on tasks such as link prediction and node classification. Unlike most approaches that represent nodes as point vectors in a low-dimensional continuous space, we embed each node as a Gaussian distribution, allowing us to capture uncertainty about the representation. Furthermore, we propose an unsupervised method that handles inductive learning scenarios and is applicable to different types of graphs: plain/attributed, directed/undirected. By leveraging both the network structure and the associated node attributes, we are able to generalize to unseen nodes without additional training. To learn the embeddings we adopt a personalized ranking formulation w.r.t. the node distances that exploits the natural ordering of the nodes imposed by the network structure. Experiments on real world networks demonstrate the high performance of our approach, outperforming state-of-the-art network embedding methods on several different tasks. Additionally, we demonstrate the benefits of modeling uncertainty - by analyzing it we can estimate neighborhood diversity and detect the intrinsic latent dimensionality of a graph.
We investigate deep generative models that can exchange multiple modalities bi-directionally, e.g., generating images from corresponding texts and vice versa. A major approach to achieve this objective is to train a model that integrates all the information of different modalities into a joint representation and then to generate one modality from the corresponding other modality via this joint representation. We simply applied this approach to variational autoencoders (VAEs), which we call a joint multimodal variational autoencoder (JMVAE). However, we found that when this model attempts to generate a large dimensional modality missing at the input, the joint representation collapses and this modality cannot be generated successfully. Furthermore, we confirmed that this difficulty cannot be resolved even using a known solution. Therefore, in this study, we propose two models to prevent this difficulty: JMVAE-kl and JMVAE-h. Results of our experiments demonstrate that these methods can prevent the difficulty above and that they generate modalities bi-directionally with equal or higher likelihood than conventional VAE methods, which generate in only one direction. Moreover, we confirm that these methods can obtain the joint representation appropriately, so that they can generate various variations of modality by moving over the joint representation or changing the value of another modality.
We present two deep generative models based on Variational Autoencoders to improve the accuracy of drug response prediction. Our models, Perturbation Variational Autoencoder and its semi-supervised extension, Drug Response Variational Autoencoder (Dr.VAE), learn latent representation of the underlying gene states before and after drug application that depend on: (i) drug-induced biological change of each gene and (ii) overall treatment response outcome. Our VAE-based models outperform the current published benchmarks in the field by anywhere from 3 to 11% AUROC and 2 to 30% AUPR. In addition, we found that better reconstruction accuracy does not necessarily lead to improvement in classification accuracy and that jointly trained models perform better than models that minimize reconstruction error independently.