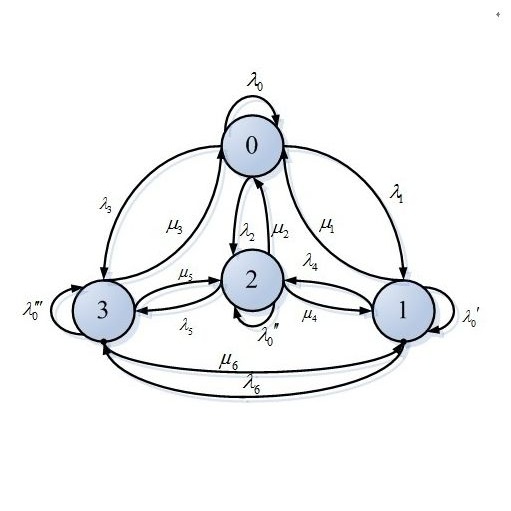
The logistic and probit link functions are the most common choices for regression models with a binary response. However, these choices are not robust to the presence of outliers/unexpected observations. The robit link function, which is equal to the inverse CDF of the Student's $t$-distribution, provides a robust alternative to the probit and logistic link functions. A multivariate normal prior for the regression coefficients is the standard choice for Bayesian inference in robit regression models. The resulting posterior density is intractable and a Data Augmentation (DA) Markov chain is used to generate approximate samples from the desired posterior distribution. Establishing geometric ergodicity for this DA Markov chain is important as it provides theoretical guarantees for asymptotic validity of MCMC standard errors for desired posterior expectations/quantiles. Previous work [Roy(2012)] established geometric ergodicity of this robit DA Markov chain assuming (i) the sample size $n$ dominates the number of predictors $p$, and (ii) an additional constraint which requires the sample size to be bounded above by a fixed constant which depends on the design matrix $X$. In particular, modern high-dimensional settings where $n < p$ are not considered. In this work, we show that the robit DA Markov chain is trace-class (i.e., the eigenvalues of the corresponding Markov operator are summable) for arbitrary choices of the sample size $n$, the number of predictors $p$, the design matrix $X$, and the prior mean and variance parameters. The trace-class property implies geometric ergodicity. Moreover, this property allows us to conclude that the sandwich robit chain (obtained by inserting an inexpensive extra step in between the two steps of the DA chain) is strictly better than the robit DA chain in an appropriate sense.
相關內容
The manuscript discusses how to incorporate random effects for quantile regression models for clustered data with focus on settings with many but small clusters. The paper has three contributions: (i) documenting that existing methods may lead to severely biased estimators for fixed effects parameters; (ii) proposing a new two-step estimation methodology where predictions of the random effects are first computed {by a pseudo likelihood approach (the LQMM method)} and then used as offsets in standard quantile regression; (iii) proposing a novel bootstrap sampling procedure in order to reduce bias of the two-step estimator and compute confidence intervals. The proposed estimation and associated inference is assessed numerically through rigorous simulation studies and applied to an AIDS Clinical Trial Group (ACTG) study.
We establish statistical properties of random-weighting methods in LASSO regression under different regularization parameters $\lambda_n$ and suitable regularity conditions. The random-weighting methods in view concern repeated optimization of a randomized objective function, motivated by the need for computational approximations to Bayesian posterior sampling. In the context of LASSO regression, we repeatedly assign analyst-drawn random weights to terms in the objective function (including the penalty terms), and optimize to obtain a sample of random-weighting estimators. We show that existing approaches have conditional model selection consistency and conditional asymptotic normality at different growth rates of $\lambda_n$ as $n \to \infty$. We propose an extension to the available random-weighting methods and establish that the resulting samples attain conditional sparse normality and conditional consistency in a growing-dimension setting. We find that random-weighting has both approximate-Bayesian and sampling-theory interpretations. Finally, we illustrate the proposed methodology via extensive simulation studies and a benchmark data example.
Overparametrized neural networks tend to perfectly fit noisy training data yet generalize well on test data. Inspired by this empirical observation, recent work has sought to understand this phenomenon of benign overfitting or harmless interpolation in the much simpler linear model. Previous theoretical work critically assumes that either the data features are statistically independent or the input data is high-dimensional; this precludes general nonparametric settings with structured feature maps. In this paper, we present a general and flexible framework for upper bounding regression and classification risk in a reproducing kernel Hilbert space. A key contribution is that our framework describes precise sufficient conditions on the data Gram matrix under which harmless interpolation occurs. Our results recover prior independent-features results (with a much simpler analysis), but they furthermore show that harmless interpolation can occur in more general settings such as features that are a bounded orthonormal system. Furthermore, our results show an asymptotic separation between classification and regression performance in a manner that was previously only shown for Gaussian features.
This paper studies the problem of statistical inference for genetic relatedness between binary traits based on individual-level genome-wide association data. Specifically, under the high-dimensional logistic regression model, we define parameters characterizing the cross-trait genetic correlation, the genetic covariance and the trait-specific genetic variance. A novel weighted debiasing method is developed for the logistic Lasso estimator and computationally efficient debiased estimators are proposed. The rates of convergence for these estimators are studied and their asymptotic normality is established under mild conditions. Moreover, we construct confidence intervals and statistical tests for these parameters, and provide theoretical justifications for the methods, including the coverage probability and expected length of the confidence intervals, as well as the size and power of the proposed tests. Numerical studies are conducted under both model generated data and simulated genetic data to show the superiority of the proposed methods and their applicability to the analysis of real genetic data. Finally, by analyzing a real data set on autoimmune diseases, we demonstrate the ability to obtain novel insights about the shared genetic architecture between ten pediatric autoimmune diseases.
The underlying physics of astronomical systems governs the relation between their measurable properties. Consequently, quantifying the statistical relationships between system-level observable properties of a population offers insights into the astrophysical drivers of that class of systems. While purely linear models capture behavior over a limited range of system scale, the fact that astrophysics is ultimately scale-dependent implies the need for a more flexible approach to describing population statistics over a wide dynamic range. For such applications, we introduce and implement a class of Kernel-Localized Linear Regression (KLLR) models. KLLR is a natural extension to the commonly-used linear models that allows the parameters of the linear model -- normalization, slope, and covariance matrix -- to be scale-dependent. KLLR performs inference in two steps: (1) it estimates the mean relation between a set of independent variables and a dependent variable and; (2) it estimates the conditional covariance of the dependent variables given a set of independent variables. We demonstrate the model's performance in a simulated setting and showcase an application of the proposed model in analyzing the baryonic content of dark matter halos. As a part of this work, we publicly release a Python implementation of the KLLR method.
We study the theoretical properties of the fused lasso procedure originally proposed by \cite{tibshirani2005sparsity} in the context of a linear regression model in which the regression coefficient are totally ordered and assumed to be sparse and piecewise constant. Despite its popularity, to the best of our knowledge, estimation error bounds in high-dimensional settings have only been obtained for the simple case in which the design matrix is the identity matrix. We formulate a novel restricted isometry condition on the design matrix that is tailored to the fused lasso estimator and derive estimation bounds for both the constrained version of the fused lasso assuming dense coefficients and for its penalised version. We observe that the estimation error can be dominated by either the lasso or the fused lasso rate, depending on whether the number of non-zero coefficient is larger than the number of piece-wise constant segments. Finally, we devise a post-processing procedure to recover the piecewise-constant pattern of the coefficients. Extensive numerical experiments support our theoretical findings.
We provide results that exactly quantify how data augmentation affects the convergence rate and variance of estimates. They lead to some unexpected findings: Contrary to common intuition, data augmentation may increase rather than decrease uncertainty of estimates, such as the empirical prediction risk. Our main theoretical tool is a limit theorem for functions of randomly transformed, high-dimensional random vectors. The proof draws on work in probability on noise stability of functions of many variables. The pathological behavior we identify is not a consequence of complex models, but can occur even in the simplest settings -- one of our examples is a linear ridge regressor with two parameters. On the other hand, our results also show that data augmentation can have real, quantifiable benefits.
We study the problem of learning causal models from observational data through the lens of interpolation and its counterpart -- regularization. A large volume of recent theoretical, as well as empirical work, suggests that, in highly complex model classes, interpolating estimators can have good statistical generalization properties and can even be optimal for statistical learning. Motivated by an analogy between statistical and causal learning recently highlighted by Janzing (2019), we investigate whether interpolating estimators can also learn good causal models. To this end, we consider a simple linearly confounded model and derive precise asymptotics for the *causal risk* of the min-norm interpolator and ridge-regularized regressors in the high-dimensional regime. Under the principle of independent causal mechanisms, a standard assumption in causal learning, we find that interpolators cannot be optimal and causal learning requires stronger regularization than statistical learning. This resolves a recent conjecture in Janzing (2019). Beyond this assumption, we find a larger range of behavior that can be precisely characterized with a new measure of *confounding strength*. If the confounding strength is negative, causal learning requires weaker regularization than statistical learning, interpolators can be optimal, and the optimal regularization can even be negative. If the confounding strength is large, the optimal regularization is infinite, and learning from observational data is actively harmful.
Ensemble methods based on subsampling, such as random forests, are popular in applications due to their high predictive accuracy. Existing literature views a random forest prediction as an infinite-order incomplete U-statistic to quantify its uncertainty. However, these methods focus on a small subsampling size of each tree, which is theoretically valid but practically limited. This paper develops an unbiased variance estimator based on incomplete U-statistics, which allows the tree size to be comparable with the overall sample size, making statistical inference possible in a broader range of real applications. Simulation results demonstrate that our estimators enjoy lower bias and more accurate confidence interval coverage without additional computational costs. We also propose a local smoothing procedure to reduce the variation of our estimator, which shows improved numerical performance when the number of trees is relatively small. Further, we investigate the ratio consistency of our proposed variance estimator under specific scenarios. In particular, we develop a new "double U-statistic" formulation to analyze the Hoeffding decomposition of the estimator's variance.
Residual networks (ResNets) have displayed impressive results in pattern recognition and, recently, have garnered considerable theoretical interest due to a perceived link with neural ordinary differential equations (neural ODEs). This link relies on the convergence of network weights to a smooth function as the number of layers increases. We investigate the properties of weights trained by stochastic gradient descent and their scaling with network depth through detailed numerical experiments. We observe the existence of scaling regimes markedly different from those assumed in neural ODE literature. Depending on certain features of the network architecture, such as the smoothness of the activation function, one may obtain an alternative ODE limit, a stochastic differential equation or neither of these. These findings cast doubts on the validity of the neural ODE model as an adequate asymptotic description of deep ResNets and point to an alternative class of differential equations as a better description of the deep network limit.