In this work, we present a deep learning framework for multi-class breast cancer image classification as our submission to the International Conference on Image Analysis and Recognition (ICIAR) 2018 Grand Challenge on BreAst Cancer Histology images (BACH). As these histology images are too large to fit into GPU memory, we first propose using Inception V3 to perform patch level classification. The patch level predictions are then passed through an ensemble fusion framework involving majority voting, gradient boosting machine (GBM), and logistic regression to obtain the image level prediction. We improve the sensitivity of the Normal and Benign predicted classes by designing a Dual Path Network (DPN) to be used as a feature extractor where these extracted features are further sent to a second layer of ensemble prediction fusion using GBM, logistic regression, and support vector machine (SVM) to refine predictions. Experimental results demonstrate our framework shows a 12.5$\%$ improvement over the state-of-the-art model.
相關內容
Few-shot image classification aims to classify unseen classes with limited labeled samples. Recent works benefit from the meta-learning process with episodic tasks and can fast adapt to class from training to testing. Due to the limited number of samples for each task, the initial embedding network for meta learning becomes an essential component and can largely affects the performance in practice. To this end, many pre-trained methods have been proposed, and most of them are trained in supervised way with limited transfer ability for unseen classes. In this paper, we proposed to train a more generalized embedding network with self-supervised learning (SSL) which can provide slow and robust representation for downstream tasks by learning from the data itself. We evaluate our work by extensive comparisons with previous baseline methods on two few-shot classification datasets ({\em i.e.,} MiniImageNet and CUB). Based on the evaluation results, the proposed method achieves significantly better performance, i.e., improve 1-shot and 5-shot tasks by nearly \textbf{3\%} and \textbf{4\%} on MiniImageNet, by nearly \textbf{9\%} and \textbf{3\%} on CUB. Moreover, the proposed method can gain the improvement of (\textbf{15\%}, \textbf{13\%}) on MiniImageNet and (\textbf{15\%}, \textbf{8\%}) on CUB by pretraining using more unlabeled data. Our code will be available at \hyperref[//github.com/phecy/SSL-FEW-SHOT.]{//github.com/phecy/ssl-few-shot.}
Recently, label consistent k-svd(LC-KSVD) algorithm has been successfully applied in image classification. The objective function of LC-KSVD is consisted of reconstruction error, classification error and discriminative sparse codes error with l0-norm sparse regularization term. The l0-norm, however, leads to NP-hard issue. Despite some methods such as orthogonal matching pursuit can help solve this problem to some extent, it is quite difficult to find the optimum sparse solution. To overcome this limitation, we propose a label embedded dictionary learning(LEDL) method to utilise the $\ell_1$-norm as the sparse regularization term so that we can avoid the hard-to-optimize problem by solving the convex optimization problem. Alternating direction method of multipliers and blockwise coordinate descent algorithm are then used to optimize the corresponding objective function. Extensive experimental results on six benchmark datasets illustrate that the proposed algorithm has achieved superior performance compared to some conventional classification algorithms.
3D image segmentation plays an important role in biomedical image analysis. Many 2D and 3D deep learning models have achieved state-of-the-art segmentation performance on 3D biomedical image datasets. Yet, 2D and 3D models have their own strengths and weaknesses, and by unifying them together, one may be able to achieve more accurate results. In this paper, we propose a new ensemble learning framework for 3D biomedical image segmentation that combines the merits of 2D and 3D models. First, we develop a fully convolutional network based meta-learner to learn how to improve the results from 2D and 3D models (base-learners). Then, to minimize over-fitting for our sophisticated meta-learner, we devise a new training method that uses the results of the base-learners as multiple versions of "ground truths". Furthermore, since our new meta-learner training scheme does not depend on manual annotation, it can utilize abundant unlabeled 3D image data to further improve the model. Extensive experiments on two public datasets (the HVSMR 2016 Challenge dataset and the mouse piriform cortex dataset) show that our approach is effective under fully-supervised, semi-supervised, and transductive settings, and attains superior performance over state-of-the-art image segmentation methods.
Deep learning has shown promising results in medical image analysis, however, the lack of very large annotated datasets confines its full potential. Although transfer learning with ImageNet pre-trained classification models can alleviate the problem, constrained image sizes and model complexities can lead to unnecessary increase in computational cost and decrease in performance. As many common morphological features are usually shared by different classification tasks of an organ, it is greatly beneficial if we can extract such features to improve classification with limited samples. Therefore, inspired by the idea of curriculum learning, we propose a strategy for building medical image classifiers using features from segmentation networks. By using a segmentation network pre-trained on similar data as the classification task, the machine can first learn the simpler shape and structural concepts before tackling the actual classification problem which usually involves more complicated concepts. Using our proposed framework on a 3D three-class brain tumor type classification problem, we achieved 82% accuracy on 191 testing samples with 91 training samples. When applying to a 2D nine-class cardiac semantic level classification problem, we achieved 86% accuracy on 263 testing samples with 108 training samples. Comparisons with ImageNet pre-trained classifiers and classifiers trained from scratch are presented.
Data augmentation has been widely used for training deep learning systems for medical image segmentation and plays an important role in obtaining robust and transformation-invariant predictions. However, it has seldom been used at test time for segmentation and not been formulated in a consistent mathematical framework. In this paper, we first propose a theoretical formulation of test-time augmentation for deep learning in image recognition, where the prediction is obtained through estimating its expectation by Monte Carlo simulation with prior distributions of parameters in an image acquisition model that involves image transformations and noise. We then propose a novel uncertainty estimation method based on the formulated test-time augmentation. Experiments with segmentation of fetal brains and brain tumors from 2D and 3D Magnetic Resonance Images (MRI) showed that 1) our test-time augmentation outperforms a single-prediction baseline and dropout-based multiple predictions, and 2) it provides a better uncertainty estimation than calculating the model-based uncertainty alone and helps to reduce overconfident incorrect predictions.
In this paper, we focus on three problems in deep learning based medical image segmentation. Firstly, U-net, as a popular model for medical image segmentation, is difficult to train when convolutional layers increase even though a deeper network usually has a better generalization ability because of more learnable parameters. Secondly, the exponential ReLU (ELU), as an alternative of ReLU, is not much different from ReLU when the network of interest gets deep. Thirdly, the Dice loss, as one of the pervasive loss functions for medical image segmentation, is not effective when the prediction is close to ground truth and will cause oscillation during training. To address the aforementioned three problems, we propose and validate a deeper network that can fit medical image datasets that are usually small in the sample size. Meanwhile, we propose a new loss function to accelerate the learning process and a combination of different activation functions to improve the network performance. Our experimental results suggest that our network is comparable or superior to state-of-the-art methods.
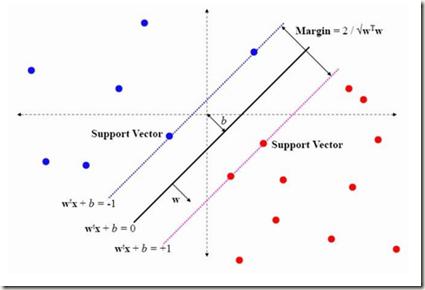
Clustering and classification critically rely on distance metrics that provide meaningful comparisons between data points. We present mixed-integer optimization approaches to find optimal distance metrics that generalize the Mahalanobis metric extensively studied in the literature. Additionally, we generalize and improve upon leading methods by removing reliance on pre-designated "target neighbors," "triplets," and "similarity pairs." Another salient feature of our method is its ability to enable active learning by recommending precise regions to sample after an optimal metric is computed to improve classification performance. This targeted acquisition can significantly reduce computational burden by ensuring training data completeness, representativeness, and economy. We demonstrate classification and computational performance of the algorithms through several simple and intuitive examples, followed by results on real image and medical datasets.
In the last years, neural networks have proven to be a powerful framework for various image analysis problems. However, some application domains have specific limitations. Notably, digital pathology is an example of such fields due to tremendous image sizes and quite limited number of training examples available. In this paper, we adopt state-of-the-art convolutional neural networks (CNN) architectures for digital pathology images analysis. We propose to classify image patches to increase effective sample size and then to apply an ensembling technique to build prediction for the original images. To validate the developed approaches, we conducted experiments with \textit{Breast Cancer Histology Challenge} dataset and obtained 90\% accuracy for the 4-class tissue classification task.
We propose an Active Learning approach to image segmentation that exploits geometric priors to streamline the annotation process. We demonstrate this for both background-foreground and multi-class segmentation tasks in 2D images and 3D image volumes. Our approach combines geometric smoothness priors in the image space with more traditional uncertainty measures to estimate which pixels or voxels are most in need of annotation. For multi-class settings, we additionally introduce two novel criteria for uncertainty. In the 3D case, we use the resulting uncertainty measure to show the annotator voxels lying on the same planar patch, which makes batch annotation much easier than if they were randomly distributed in the volume. The planar patch is found using a branch-and-bound algorithm that finds a patch with the most informative instances. We evaluate our approach on Electron Microscopy and Magnetic Resonance image volumes, as well as on regular images of horses and faces. We demonstrate a substantial performance increase over state-of-the-art approaches.
While deep convolutional neural networks (CNNs) have shown a great success in single-label image classification, it is important to note that real world images generally contain multiple labels, which could correspond to different objects, scenes, actions and attributes in an image. Traditional approaches to multi-label image classification learn independent classifiers for each category and employ ranking or thresholding on the classification results. These techniques, although working well, fail to explicitly exploit the label dependencies in an image. In this paper, we utilize recurrent neural networks (RNNs) to address this problem. Combined with CNNs, the proposed CNN-RNN framework learns a joint image-label embedding to characterize the semantic label dependency as well as the image-label relevance, and it can be trained end-to-end from scratch to integrate both information in a unified framework. Experimental results on public benchmark datasets demonstrate that the proposed architecture achieves better performance than the state-of-the-art multi-label classification model