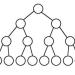
A permutation graph can be defined as an intersection graph of segments whose endpoints lie on two parallel lines $\ell_1$ and $\ell_2$, one on each. A bipartite permutation graph is a permutation graph which is bipartite. In the the bipartite permutation vertex deletion problem we ask for a given $n$-vertex graph, whether we can remove at most $k$ vertices to obtain a bipartite permutation graph. This problem is NP-complete but it does admit an FPT algorithm parameterized by $k$. In this paper we study the kernelization of this problem and show that it admits a polynomial kernel with $O(k^{99})$ vertices.
相關內容
A long line of research on fixed parameter tractability of integer programming culminated with showing that integer programs with n variables and a constraint matrix with dual tree-depth d and largest entry D are solvable in time g(d,D)poly(n) for some function g. However, the dual tree-depth of a constraint matrix is not preserved by row operations, i.e., a given integer program can be equivalent to another with a smaller dual tree-depth, and thus does not reflect its geometric structure. We prove that the minimum dual tree-depth of a row-equivalent matrix is equal to the branch-depth of the matroid defined by the columns of the matrix. We design a fixed parameter algorithm for computing branch-depth of matroids represented over a finite field and a fixed parameter algorithm for computing a row-equivalent matrix with minimum dual tree-depth. Finally, we use these results to obtain an algorithm for integer programming running in time g(d*,D)poly(n) where d* is the branch-depth of the constraint matrix; the branch-depth cannot be replaced by the more permissive notion of branch-width.
We consider edge modification problems towards block and strictly chordal graphs, where one is given an undirected graph $G = (V,E)$ and an integer $k \in \mathbb{N}$ and seeks to edit (add or delete) at most $k$ edges from $G$ to obtain a block graph or a strictly chordal graph. The completion and deletion variants of these problems are defined similarly by only allowing edge additions for the former and only edge deletions for the latter. Block graphs are a well-studied class of graphs and admit several characterizations, e.g. they are diamond-free chordal graphs. Strictly chordal graphs, also referred to as block duplicate graphs, are a natural generalization of block graphs where one can add true twins of cut-vertices. Strictly chordal graphs are exactly dart and gem-free chordal graphs. We prove the NP-completeness for most variants of these problems and provide $O(k^2)$ vertex-kernels for Block Graph Edition and Block Graph Deletion, $O(k^3)$ vertex-kernels for Strictly Chordal Completion and Strictly Chordal Deletion and a $O(k^4)$ vertex-kernel for Strictly Chordal Edition.
In this paper, we show several parameterized problems to be complete for the class XNLP: parameterized problems that can be solved with a non-deterministic algorithm that uses $f(k)\log n$ space and $f(k)n^c$ time, with $f$ a computable function, $n$ the input size, $k$ the parameter and $c$ a constant. The problems include Maximum Regular Induced Subgraph and Max Cut parameterized by linear clique-width, Capacitated (Red-Blue) Dominating Set parameterized by pathwidth, Odd Cycle Transversal parameterized by a new parameter we call logarithmic linear clique-width (defined as $k/\log n$ for an $n$-vertex graph of linear clique-width $k$), and Bipartite Bandwidth.
Let $G=(V(G),E(G))$ be a finite simple undirected graph with vertex set $V(G)$, edge set $E(G)$ and vertex subset $S\subseteq V(G)$. $S$ is termed \emph{open-dominating} if every vertex of $G$ has at least one neighbor in $S$, and \emph{open-independent, open-locating-dominating} (an $OLD_{oind}$-set for short) if no two vertices in $G$ have the same set of neighbors in $S$, and each vertex in $S$ is open-dominated exactly once by $S$. The problem of deciding whether or not $G$ has an $OLD_{oind}$-set has important applications that have been reported elsewhere. As the problem is known to be $\mathcal{NP}$-complete, it appears to be notoriously difficult as we show that its complexity remains the same even for just planar bipartite graphs of maximum degree five and girth six, and also for planar subcubic graphs of girth nine. Also, we present characterizations of both $P_4$-tidy graphs and the complementary prisms of cographs that have an $OLD_{oind}$-set.
We study the problem of computing the vitality with respect to max flow of edges and vertices in undirected planar graphs, where the vitality of an edge/vertex in a graph with respect to max flow between two fixed vertices $s,t$ is defined as the max flow decrease when the edge/vertex is removed from the graph. We show that the vitality of any $k$ selected edges can be computed in $O(kn + n\log\log n)$ worst-case time, and that a $\delta$ additive approximation of the vitality of all edges with capacity at most $c$ can be computed in $O(\frac{c}{\delta}n +n \log \log n)$ worst-case time, where $n$ is the size of the graph. Similar results are given for the vitality of vertices. All our algorithms work in $O(n)$ space.
We contribute to fulfil the long-lasting gap in the understanding of the spatial search with multiple marked vertices. The theoretical framework is that of discrete-time quantum walks (QW), i.e. local unitary matrices that drive the evolution of a single particle on the lattice. QW based search algorithms are well understood when they have to tackle the fundamental problem of finding only one marked element in a $d-$dimensional grid and it has been proven they provide a quadratic advantage over classical searching protocols. However, once we consider to search more than one element, the behaviour of the algorithm may be affected by the spatial configuration of the marked elements, due to the quantum interference among themselves and even the quantum advantage is no longer granted. Here our main contribution is twofold : (i) we provide strong numerical evidence that spatial configurations are almost all optimal; and (ii) we analytically prove that the quantum advantage with respect to the classical counterpart is not always granted and it does depend on the proportion of searched elements over the total number of grid points $\tau$. We finally providing a clear phase diagram for the QW search advantage with respect to the classical random algorithm.
Zero-free based algorithm is a major technique for deterministic approximate counting. In Barvinok's original framework[Bar17], by calculating truncated Taylor expansions, a quasi-polynomial time algorithm was given for estimating zero-free partition functions. Patel and Regts[PR17] later gave a refinement of Barvinok's framework, which gave a polynomial-time algorithm for a class of zero-free graph polynomials that can be expressed as counting induced subgraphs in bounded-degree graphs. In this paper, we give a polynomial-time algorithm for estimating classical and quantum partition functions specified by local Hamiltonians with bounded maximum degree, assuming a zero-free property for the temperature. Consequently, when the inverse temperature is close enough to zero by a constant gap, we have polynomial-time approximation algorithm for all such partition functions. Our result is based on a new abstract framework that extends and generalizes the approach of Patel and Regts.
A typical task in temporal graphs analysis is answering single-source-all-destination (SSAD) temporal distance queries. An SSAD query starting at a vertex $v$ asks for the temporal distances, e.g., durations or earliest arrival times between $v$ and all other vertices. We introduce an index to speed up SSAD temporal distance queries called Substream index. The indexing is based on the construction of $k$ subgraphs and a mapping from the vertices to the subgraphs. Each subgraph contains the temporal edges sufficient to answer queries starting from any vertex mapped to the subgraph. We answer a query starting at a vertex $v$ with a single pass over the edges of the subgraph. Our index supports dynamic updates, i.e., efficient insertion and deletion of temporal edges. Unfortunately, deciding if a Substream index of a given size exists is NP-complete. However, we provide an efficient greedy approximation that constructs an index at most $k/\delta$ times larger than an optimal index where $\delta$, with $1\leq\delta\leq k$, depends on the temporal and spatial structure of the graph. Moreover, we improve the running time of the approximation in three ways. First, we use an auxiliary index called Time Skip index to speed up the construction and queries by skipping edges that do not need to be considered. Next, we apply min-hashing to avoid costly union operations. Finally, we use parallelization to take the parallel processing capabilities of modern processors into account. An extensive evaluation using real-world temporal networks shows the efficiency and effectiveness of our indices, and query times are significantly improved for all data sets.
Aligning a sequence to a walk in a labeled graph is a problem of fundamental importance to Computational Biology. For finding a walk in an arbitrary graph with $|E|$ edges that exactly matches a pattern of length $m$, a lower bound based on the Strong Exponential Time Hypothesis (SETH) implies an algorithm significantly faster than $O(|E|m)$ time is unlikely [Equi et al., ICALP 2019]. However, for many special graphs, such as de Bruijn graphs, the problem can be solved in linear time [Bowe et al., WABI 2012]. For approximate matching, the picture is more complex. When edits (substitutions, insertions, and deletions) are only allowed to the pattern, or when the graph is acyclic, the problem is again solvable in $O(|E|m)$ time. When edits are allowed to arbitrary cyclic graphs, the problem becomes NP-complete, even on binary alphabets [Jain et al., RECOMB 2019]. These results hold even when edits are restricted to only substitutions. The complexity of approximate pattern matching on de Bruijn graphs remained open. We investigate this problem and show that the properties that make de Bruijn graphs amenable to efficient exact pattern matching do not extend to approximate matching, even when restricted to the substitutions only case with alphabet size four. We prove that determining the existence of a matching walk in a de Bruijn graph is NP-complete when substitutions are allowed to the graph. In addition, we demonstrate that an algorithm significantly faster than $O(|E|m)$ is unlikely for de Bruijn graphs in the case where only substitutions are allowed to the pattern. This stands in contrast to pattern-to-text matching where exact matching is solvable in linear time, like on de Bruijn graphs, but approximate matching under substitutions is solvable in subquadratic $O(n\sqrt{m})$ time, where $n$ is the text's length [Abrahamson, SIAM J. Computing 1987].
The median of a graph $G$ with weighted vertices is the set of all vertices $x$ minimizing the sum of weighted distances from $x$ to the vertices of $G$. For any integer $p\ge 2$, we characterize the graphs in which, with respect to any non-negative weights, median sets always induce connected subgraphs in the $p$th power $G^p$ of $G$. This extends some characterizations of graphs with connected medians (case $p=1$) provided by Bandelt and Chepoi (2002). The characteristic conditions can be tested in polynomial time for any $p$. We also show that several important classes of graphs in metric graph theory, including bridged graphs (and thus chordal graphs), graphs with convex balls, bucolic graphs, and bipartite absolute retracts, have $G^2$-connected medians. Extending the result of Bandelt and Chepoi that basis graphs of matroids are graphs with connected medians, we characterize the isometric subgraphs of Johnson graphs and of halved-cubes with connected medians.