Medulloblastoma (MB) is a primary central nervous system tumor and the most common malignant brain cancer among children. Neuropathologists perform microscopic inspection of histopathological tissue slides under a microscope to assess the severity of the tumor. This is a time-consuming task and often infused with observer variability. Recently, pre-trained convolutional neural networks (CNN) have shown promising results for MB subtype classification. Typically, high-resolution images are divided into smaller tiles for classification, while the size of the tiles has not been systematically evaluated. We study the impact of tile size and input strategy and classify the two major histopathological subtypes-Classic and Demoplastic/Nodular. To this end, we use recently proposed EfficientNets and evaluate tiles with increasing size combined with various downsampling scales. Our results demonstrate using large input tiles pixels followed by intermediate downsampling and patch cropping significantly improves MB classification performance. Our top-performing method achieves the AUC-ROC value of 90.90\% compared to 84.53\% using the previous approach with smaller input tiles.
相關內容
Image segmentation is a key topic in image processing and computer vision with applications such as scene understanding, medical image analysis, robotic perception, video surveillance, augmented reality, and image compression, among many others. Various algorithms for image segmentation have been developed in the literature. Recently, due to the success of deep learning models in a wide range of vision applications, there has been a substantial amount of works aimed at developing image segmentation approaches using deep learning models. In this survey, we provide a comprehensive review of the literature at the time of this writing, covering a broad spectrum of pioneering works for semantic and instance-level segmentation, including fully convolutional pixel-labeling networks, encoder-decoder architectures, multi-scale and pyramid based approaches, recurrent networks, visual attention models, and generative models in adversarial settings. We investigate the similarity, strengths and challenges of these deep learning models, examine the most widely used datasets, report performances, and discuss promising future research directions in this area.
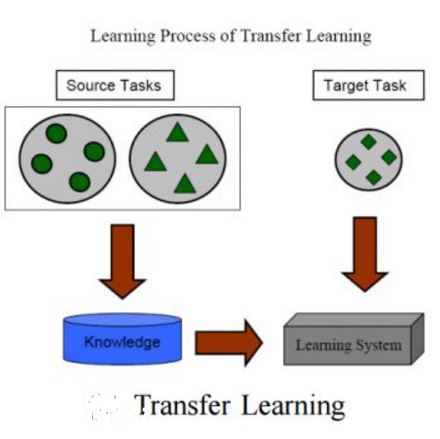
Meta-learning has been proposed as a framework to address the challenging few-shot learning setting. The key idea is to leverage a large number of similar few-shot tasks in order to learn how to adapt a base-learner to a new task for which only a few labeled samples are available. As deep neural networks (DNNs) tend to overfit using a few samples only, meta-learning typically uses shallow neural networks (SNNs), thus limiting its effectiveness. In this paper we propose a novel few-shot learning method called meta-transfer learning (MTL) which learns to adapt a deep NN for few shot learning tasks. Specifically, "meta" refers to training multiple tasks, and "transfer" is achieved by learning scaling and shifting functions of DNN weights for each task. In addition, we introduce the hard task (HT) meta-batch scheme as an effective learning curriculum for MTL. We conduct experiments using (5-class, 1-shot) and (5-class, 5-shot) recognition tasks on two challenging few-shot learning benchmarks: miniImageNet and Fewshot-CIFAR100. Extensive comparisons to related works validate that our meta-transfer learning approach trained with the proposed HT meta-batch scheme achieves top performance. An ablation study also shows that both components contribute to fast convergence and high accuracy.
Medical image segmentation is a primary task in many applications, and the accuracy of the segmentation is a necessity. Recently, many deep learning networks derived from U-Net have been extensively used and have achieved notable results. To further improve and refine the performance of U-Net, parallel decoders along with mask prediction decoder have been carried out and have shown significant improvement with additional advantages. In our work, we utilize the advantages of using a combination of contour and distance map as regularizers. In turn, we propose a novel architecture Psi-Net with a single encoder and three parallel decoders, one decoder to learn the mask and other two to learn the auxiliary tasks of contour detection and distance map estimation. The learning of these auxiliary tasks helps in capturing the shape and boundary. We also propose a new joint loss function for the proposed architecture. The loss function consists of a weighted combination of Negative likelihood and Mean Square Error loss. We have used two publicly available datasets: 1) Origa dataset for the task of optic cup and disc segmentation and 2) Endovis segment dataset for the task of polyp segmentation to evaluate our model. We have conducted extensive experiments using our network to show our model gives better results in terms of segmentation, boundary and shape metrics.
3D image segmentation plays an important role in biomedical image analysis. Many 2D and 3D deep learning models have achieved state-of-the-art segmentation performance on 3D biomedical image datasets. Yet, 2D and 3D models have their own strengths and weaknesses, and by unifying them together, one may be able to achieve more accurate results. In this paper, we propose a new ensemble learning framework for 3D biomedical image segmentation that combines the merits of 2D and 3D models. First, we develop a fully convolutional network based meta-learner to learn how to improve the results from 2D and 3D models (base-learners). Then, to minimize over-fitting for our sophisticated meta-learner, we devise a new training method that uses the results of the base-learners as multiple versions of "ground truths". Furthermore, since our new meta-learner training scheme does not depend on manual annotation, it can utilize abundant unlabeled 3D image data to further improve the model. Extensive experiments on two public datasets (the HVSMR 2016 Challenge dataset and the mouse piriform cortex dataset) show that our approach is effective under fully-supervised, semi-supervised, and transductive settings, and attains superior performance over state-of-the-art image segmentation methods.
We address the problem of segmenting 3D multi-modal medical images in scenarios where very few labeled examples are available for training. Leveraging the recent success of adversarial learning for semi-supervised segmentation, we propose a novel method based on Generative Adversarial Networks (GANs) to train a segmentation model with both labeled and unlabeled images. The proposed method prevents over-fitting by learning to discriminate between true and fake patches obtained by a generator network. Our work extends current adversarial learning approaches, which focus on 2D single-modality images, to the more challenging context of 3D volumes of multiple modalities. The proposed method is evaluated on the problem of segmenting brain MRI from the iSEG-2017 and MRBrainS 2013 datasets. Significant performance improvement is reported, compared to state-of-art segmentation networks trained in a fully-supervised manner. In addition, our work presents a comprehensive analysis of different GAN architectures for semi-supervised segmentation, showing recent techniques like feature matching to yield a higher performance than conventional adversarial training approaches. Our code is publicly available at //github.com/arnab39/FewShot_GAN-Unet3D
Deep learning has shown promising results in medical image analysis, however, the lack of very large annotated datasets confines its full potential. Although transfer learning with ImageNet pre-trained classification models can alleviate the problem, constrained image sizes and model complexities can lead to unnecessary increase in computational cost and decrease in performance. As many common morphological features are usually shared by different classification tasks of an organ, it is greatly beneficial if we can extract such features to improve classification with limited samples. Therefore, inspired by the idea of curriculum learning, we propose a strategy for building medical image classifiers using features from segmentation networks. By using a segmentation network pre-trained on similar data as the classification task, the machine can first learn the simpler shape and structural concepts before tackling the actual classification problem which usually involves more complicated concepts. Using our proposed framework on a 3D three-class brain tumor type classification problem, we achieved 82% accuracy on 191 testing samples with 91 training samples. When applying to a 2D nine-class cardiac semantic level classification problem, we achieved 86% accuracy on 263 testing samples with 108 training samples. Comparisons with ImageNet pre-trained classifiers and classifiers trained from scratch are presented.
Over the past decades, state-of-the-art medical image segmentation has heavily rested on signal processing paradigms, most notably registration-based label propagation and pair-wise patch comparison, which are generally slow despite a high segmentation accuracy. In recent years, deep learning has revolutionalized computer vision with many practices outperforming prior art, in particular the convolutional neural network (CNN) studies on image classification. Deep CNN has also started being applied to medical image segmentation lately, but generally involves long training and demanding memory requirements, achieving limited success. We propose a patch-based deep learning framework based on a revisit to the classic neural network model with substantial modernization, including the use of Rectified Linear Unit (ReLU) activation, dropout layers, 2.5D tri-planar patch multi-pathway settings. In a test application to hippocampus segmentation using 100 brain MR images from the ADNI database, our approach significantly outperformed prior art in terms of both segmentation accuracy and speed: scoring a median Dice score up to 90.98% on a near real-time performance (<1s).
In this paper, we propose an effective electrocardiogram (ECG) arrhythmia classification method using a deep two-dimensional convolutional neural network (CNN) which recently shows outstanding performance in the field of pattern recognition. Every ECG beat was transformed into a two-dimensional grayscale image as an input data for the CNN classifier. Optimization of the proposed CNN classifier includes various deep learning techniques such as batch normalization, data augmentation, Xavier initialization, and dropout. In addition, we compared our proposed classifier with two well-known CNN models; AlexNet and VGGNet. ECG recordings from the MIT-BIH arrhythmia database were used for the evaluation of the classifier. As a result, our classifier achieved 99.05% average accuracy with 97.85% average sensitivity. To precisely validate our CNN classifier, 10-fold cross-validation was performed at the evaluation which involves every ECG recording as a test data. Our experimental results have successfully validated that the proposed CNN classifier with the transformed ECG images can achieve excellent classification accuracy without any manual pre-processing of the ECG signals such as noise filtering, feature extraction, and feature reduction.
We propose a novel locally adaptive learning estimator for enhancing the inter- and intra- discriminative capabilities of Deep Neural Networks, which can be used as improved loss layer for semantic image segmentation tasks. Most loss layers compute pixel-wise cost between feature maps and ground truths, ignoring spatial layouts and interactions between neighboring pixels with same object category, and thus networks cannot be effectively sensitive to intra-class connections. Stride by stride, our method firstly conducts adaptive pooling filter operating over predicted feature maps, aiming to merge predicted distributions over a small group of neighboring pixels with same category, and then it computes cost between the merged distribution vector and their category label. Such design can make groups of neighboring predictions from same category involved into estimations on predicting correctness with respect to their category, and hence train networks to be more sensitive to regional connections between adjacent pixels based on their categories. In the experiments on Pascal VOC 2012 segmentation datasets, the consistently improved results show that our proposed approach achieves better segmentation masks against previous counterparts.
We propose an Active Learning approach to image segmentation that exploits geometric priors to streamline the annotation process. We demonstrate this for both background-foreground and multi-class segmentation tasks in 2D images and 3D image volumes. Our approach combines geometric smoothness priors in the image space with more traditional uncertainty measures to estimate which pixels or voxels are most in need of annotation. For multi-class settings, we additionally introduce two novel criteria for uncertainty. In the 3D case, we use the resulting uncertainty measure to show the annotator voxels lying on the same planar patch, which makes batch annotation much easier than if they were randomly distributed in the volume. The planar patch is found using a branch-and-bound algorithm that finds a patch with the most informative instances. We evaluate our approach on Electron Microscopy and Magnetic Resonance image volumes, as well as on regular images of horses and faces. We demonstrate a substantial performance increase over state-of-the-art approaches.