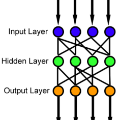
Feedforward neural networks (FNNs) can be viewed as non-linear regression models, where covariates enter the model through a combination of weighted summations and non-linear functions. Although these models have some similarities to the models typically used in statistical modelling, the majority of neural network research has been conducted outside of the field of statistics. This has resulted in a lack of statistically-based methodology, and, in particular, there has been little emphasis on model parsimony. Determining the input layer structure is analogous to variable selection, while the structure for the hidden layer relates to model complexity. In practice, neural network model selection is often carried out by comparing models using out-of-sample performance. However, in contrast, the construction of an associated likelihood function opens the door to information-criteria-based variable and architecture selection. A novel model selection method, which performs both input- and hidden-node selection, is proposed using the Bayesian information criterion (BIC) for FNNs. The choice of BIC over out-of-sample performance as the model selection objective function leads to an increased probability of recovering the true model, while parsimoniously achieving favourable out-of-sample performance. Simulation studies are used to evaluate and justify the proposed method, and applications on real data are investigated.
相關內容
Selective classification involves identifying the subset of test samples that a model can classify with high accuracy, and is important for applications such as automated medical diagnosis. We argue that this capability of identifying uncertain samples is valuable for training classifiers as well, with the aim of building more accurate classifiers. We unify these dual roles by training a single auxiliary meta-network to output an importance weight as a function of the instance. This measure is used at train time to reweight training data, and at test-time to rank test instances for selective classification. A second, key component of our proposal is the meta-objective of minimizing dropout variance (the variance of classifier output when subjected to random weight dropout) for training the metanetwork. We train the classifier together with its metanetwork using a nested objective of minimizing classifier loss on training data and meta-loss on a separate meta-training dataset. We outperform current state-of-the-art on selective classification by substantial margins--for instance, upto 1.9% AUC and 2% accuracy on a real-world diabetic retinopathy dataset. Finally, our meta-learning framework extends naturally to unsupervised domain adaptation, given our unsupervised variance minimization meta-objective. We show cumulative absolute gains of 3.4% / 3.3% accuracy and AUC over the other baselines in domain shift settings on the Retinopathy dataset using unsupervised domain adaptation.
Penalized likelihood models are widely used to simultaneously select variables and estimate model parameters. However, the existence of weak signals can lead to inaccurate variable selection, biased parameter estimation, and invalid inference. Thus, identifying weak signals accurately and making valid inferences are crucial in penalized likelihood models. We develop a unified approach to identify weak signals and make inferences in penalized likelihood models, including the special case when the responses are categorical. To identify weak signals, we use the estimated selection probability of each covariate as a measure of the signal strength and formulate a signal identification criterion. To construct confidence intervals, we propose a two-step inference procedure. Extensive simulation studies show that the proposed procedure outperforms several existing methods. We illustrate the proposed method by applying it to the Practice Fusion diabetes data set.
Active Malware Analysis involves modeling malware behavior by executing actions to trigger responses and explore multiple execution paths. One of the aims is making the action selection more efficient. This paper treats Active Malware Analysis as a Bayes-Active Markov Decision Process and uses a Bayesian Model Combination approach to train an analyzer agent. We show an improvement in performance against other Bayesian and stochastic approaches to Active Malware Analysis.
Unsupervised discretization is a crucial step in many knowledge discovery tasks. The state-of-the-art method for one-dimensional data infers locally adaptive histograms using the minimum description length (MDL) principle, but the multi-dimensional case is far less studied: current methods consider the dimensions one at a time (if not independently), which result in discretizations based on rectangular cells of adaptive size. Unfortunately, this approach is unable to adequately characterize dependencies among dimensions and/or results in discretizations consisting of more cells (or bins) than is desirable. To address this problem, we propose an expressive model class that allows for far more flexible partitions of two-dimensional data. We extend the state of the art for the one-dimensional case to obtain a model selection problem based on the normalized maximum likelihood, a form of refined MDL. As the flexibility of our model class comes at the cost of a vast search space, we introduce a heuristic algorithm, named PALM, which Partitions each dimension ALternately and then Merges neighboring regions, all using the MDL principle. Experiments on synthetic data show that PALM 1) accurately reveals ground truth partitions that are within the model class (i.e., the search space), given a large enough sample size; 2) approximates well a wide range of partitions outside the model class; 3) converges, in contrast to the state-of-the-art multivariate discretization method IPD. Finally, we apply our algorithm to three spatial datasets, and we demonstrate that, compared to kernel density estimation (KDE), our algorithm not only reveals more detailed density changes, but also fits unseen data better, as measured by the log-likelihood.
A model-based approach is developed for clustering categorical data with no natural ordering. The proposed method exploits the Hamming distance to define a family of probability mass functions to model the data. The elements of this family are then considered as kernels of a finite mixture model with unknown number of components. Conjugate Bayesian inference has been derived for the parameters of the Hamming distribution model. The mixture is framed in a Bayesian nonparametric setting and a transdimensional blocked Gibbs sampler is developed to provide full Bayesian inference on the number of clusters, their structure and the group-specific parameters, facilitating the computation with respect to customary reversible jump algorithms. The proposed model encompasses a parsimonious latent class model as a special case, when the number of components is fixed. Model performances are assessed via a simulation study and reference datasets, showing improvements in clustering recovery over existing approaches.
Mathematical models are essential for understanding and making predictions about systems arising in nature and engineering. Yet, mathematical models are a simplification of true phenomena, thus making predictions subject to uncertainty. Hence, the ability to quantify uncertainties is essential to any modelling framework, enabling the user to assess the importance of certain parameters on quantities of interest and have control over the quality of the model output by providing a rigorous understanding of uncertainty. Peridynamic models are a particular class of mathematical models that have proven to be remarkably accurate and robust for a large class of material failure problems. However, the high computational expense of peridynamic models remains a major limitation, hindering outer-loop applications that require a large number of simulations, for example, uncertainty quantification. This contribution provides a framework to make such computations feasible. By employing a Multilevel Monte Carlo (MLMC) framework, where the majority of simulations are performed using a coarse mesh, and performing relatively few simulations using a fine mesh, a significant reduction in computational cost can be realised, and statistics of structural failure can be estimated. The results show a speed-up factor of 16x over a standard Monte Carlo estimator, enabling the forward propagation of uncertain parameters in a computationally expensive peridynamic model. Furthermore, the multilevel method provides an estimate of both the discretisation error and sampling error, thus improving the confidence in numerical predictions. The performance of the approach is demonstrated through an examination of the statistical size effect in quasi-brittle materials.
Causal discovery and causal reasoning are classically treated as separate and consecutive tasks: one first infers the causal graph, and then uses it to estimate causal effects of interventions. However, such a two-stage approach is uneconomical, especially in terms of actively collected interventional data, since the causal query of interest may not require a fully-specified causal model. From a Bayesian perspective, it is also unnatural, since a causal query (e.g., the causal graph or some causal effect) can be viewed as a latent quantity subject to posterior inference -- other unobserved quantities that are not of direct interest (e.g., the full causal model) ought to be marginalized out in this process and contribute to our epistemic uncertainty. In this work, we propose Active Bayesian Causal Inference (ABCI), a fully-Bayesian active learning framework for integrated causal discovery and reasoning, which jointly infers a posterior over causal models and queries of interest. In our approach to ABCI, we focus on the class of causally-sufficient, nonlinear additive noise models, which we model using Gaussian processes. We sequentially design experiments that are maximally informative about our target causal query, collect the corresponding interventional data, and update our beliefs to choose the next experiment. Through simulations, we demonstrate that our approach is more data-efficient than several baselines that only focus on learning the full causal graph. This allows us to accurately learn downstream causal queries from fewer samples while providing well-calibrated uncertainty estimates for the quantities of interest.
In Multi-Label Text Classification (MLTC), one sample can belong to more than one class. It is observed that most MLTC tasks, there are dependencies or correlations among labels. Existing methods tend to ignore the relationship among labels. In this paper, a graph attention network-based model is proposed to capture the attentive dependency structure among the labels. The graph attention network uses a feature matrix and a correlation matrix to capture and explore the crucial dependencies between the labels and generate classifiers for the task. The generated classifiers are applied to sentence feature vectors obtained from the text feature extraction network (BiLSTM) to enable end-to-end training. Attention allows the system to assign different weights to neighbor nodes per label, thus allowing it to learn the dependencies among labels implicitly. The results of the proposed model are validated on five real-world MLTC datasets. The proposed model achieves similar or better performance compared to the previous state-of-the-art models.
Many tasks in natural language processing can be viewed as multi-label classification problems. However, most of the existing models are trained with the standard cross-entropy loss function and use a fixed prediction policy (e.g., a threshold of 0.5) for all the labels, which completely ignores the complexity and dependencies among different labels. In this paper, we propose a meta-learning method to capture these complex label dependencies. More specifically, our method utilizes a meta-learner to jointly learn the training policies and prediction policies for different labels. The training policies are then used to train the classifier with the cross-entropy loss function, and the prediction policies are further implemented for prediction. Experimental results on fine-grained entity typing and text classification demonstrate that our proposed method can obtain more accurate multi-label classification results.
With the rapid increase of large-scale, real-world datasets, it becomes critical to address the problem of long-tailed data distribution (i.e., a few classes account for most of the data, while most classes are under-represented). Existing solutions typically adopt class re-balancing strategies such as re-sampling and re-weighting based on the number of observations for each class. In this work, we argue that as the number of samples increases, the additional benefit of a newly added data point will diminish. We introduce a novel theoretical framework to measure data overlap by associating with each sample a small neighboring region rather than a single point. The effective number of samples is defined as the volume of samples and can be calculated by a simple formula $(1-\beta^{n})/(1-\beta)$, where $n$ is the number of samples and $\beta \in [0,1)$ is a hyperparameter. We design a re-weighting scheme that uses the effective number of samples for each class to re-balance the loss, thereby yielding a class-balanced loss. Comprehensive experiments are conducted on artificially induced long-tailed CIFAR datasets and large-scale datasets including ImageNet and iNaturalist. Our results show that when trained with the proposed class-balanced loss, the network is able to achieve significant performance gains on long-tailed datasets.