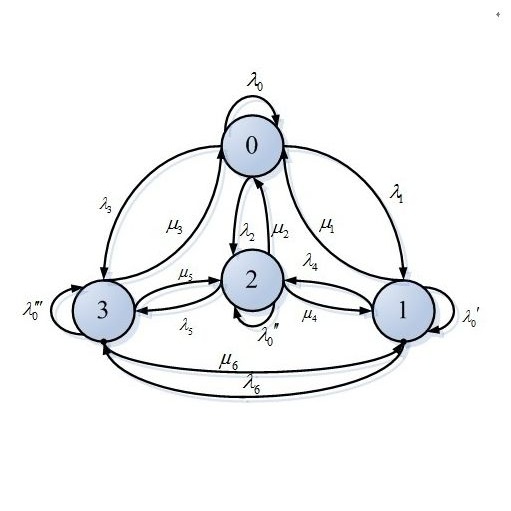
This paper presents a model for COVID19 in Mexico City. The data analyzed were considered from the appearance of the first case in Mexico until July 2021. In this first approximation the states considered were Susceptible, Infected, Hospitalized, Intensive Care Unit, Intubated, and Dead. As a consequence of the lack of coronavirus testing, the number of infected and dead people is underestimated, although the results obtained give a good approximation to the evolution of the pandemic in Mexico City. The model is based on a discrete-time Markov chain considering data provided by the Mexican government, the main objective is to estimate the transient probabilities from one state to another for the Mexico City case.
相關內容
The goals of this paper are twofold: (1) to present a new method that is able to find linear laws governing the time evolution of Markov chains and (2) to apply this method for anomaly detection in Bitcoin prices. To accomplish these goals, first, the linear laws of Markov chains are derived by using the time embedding of their (categorical) autocorrelation function. Then, a binary series is generated from the first difference of Bitcoin exchange rate (against the United States Dollar). Finally, the minimum number of parameters describing the linear laws of this series is identified through stepped time windows. Based on the results, linear laws typically became more complex (containing an additional third parameter that indicates hidden Markov property) in two periods: before the crash of cryptocurrency markets inducted by the COVID-19 pandemic (12 March 2020), and before the record-breaking surge in the price of Bitcoin (Q4 2020 - Q1 2021). In addition, the locally high values of this third parameter are often related to short-term price peaks, which suggests price manipulation.
In this paper, we use SEIR equations to make predictions for the number of mortality due to COVID-19 in \.Istanbul. Using excess mortality method, we find the number of mortality for the previous three waves in 2020 and 2021. We show that the predictions of our model is consistent with number of moralities for each wave. Furthermore, we predict the number of mortality for the second wave of 2021. We also extend our analysis for Germany, Italy and Turkey to compare the basic reproduction number $R_0$ for Istanbul. Finally, we calculate the number of infected people in Istanbul for herd immunity.
The current outbreak of a coronavirus, has quickly escalated to become a serious global problem that has now been declared a Public Health Emergency of International Concern by the World Health Organization. Infectious diseases know no borders, so when it comes to controlling outbreaks, timing is absolutely essential. It is so important to detect threats as early as possible, before they spread. After a first successful DiCOVA challenge, the organisers released second DiCOVA challenge with the aim of diagnosing COVID-19 through the use of breath, cough and speech audio samples. This work presents the details of the automatic system for COVID-19 detection using breath, cough and speech recordings. We developed different front-end auditory acoustic features along with a bidirectional Long Short-Term Memory (bi-LSTM) as classifier. The results are promising and have demonstrated the high complementary behaviour among the auditory acoustic features in the Breathing, Cough and Speech tracks giving an AUC of 86.60% on the test set.
Statistical methods are developed for analysis of clinical and virus genetics data from phase 3 randomized, placebo-controlled trials of vaccines against novel coronavirus COVID-19. Vaccine efficacy (VE) of a vaccine to prevent COVID-19 caused by one of finitely many genetic strains of SARS-CoV-2 may vary by strain. The problem of assessing differential VE by viral genetics can be formulated under a competing risks model where the endpoint is virologically confirmed COVID-19 and the cause-of-failure is the infecting SARS-CoV-2 genotype. Strain-specific VE is defined as one minus the cause-specific hazard ratio (vaccine/placebo). For the COVID-19 VE trials, the time to COVID-19 is right-censored, and a substantial percentage of failure cases are missing the infecting virus genotype. We develop estimation and hypothesis testing procedures for strain-specific VE when the failure time is subject to right censoring and the cause-of-failure is subject to missingness, focusing on $J \ge 2$ discrete categorical unordered or ordered virus genotypes. The stratified Cox proportional hazards model is used to relate the cause-specific outcomes to explanatory variables. The inverse probability weighted complete-case (IPW) estimator and the augmented inverse probability weighted complete-case (AIPW) estimator are investigated. Hypothesis tests are developed to assess whether the vaccine provides at least a specified level of efficacy against some viral genotypes and whether VE varies across genotypes, adjusting for covariates. The finite-sample properties of the proposed tests are studied through simulations and are shown to have good performances. In preparation for the real data analyses, the developed methods are applied to a pseudo dataset mimicking the Moderna COVE trial.
We study a dynamic infection spread model, inspired by the discrete time SIR model, where infections are spread via non-isolated infected individuals. While infection keeps spreading over time, a limited capacity testing is performed at each time instance as well. In contrast to the classical, static, group testing problem, the objective in our setup is not to find the minimum number of required tests to identify the infection status of every individual in the population, but to control the infection spread by detecting and isolating the infections over time by using the given, limited number of tests. In order to analyze the performance of the proposed algorithms, we focus on the mean-sense analysis of the number of individuals that remain non-infected throughout the process of controlling the infection. We propose two dynamic algorithms that both use given limited number of tests to identify and isolate the infections over time, while the infection spreads. While the first algorithm is a dynamic randomized individual testing algorithm, in the second algorithm we employ the group testing approach similar to the original work of Dorfman. By considering weak versions of our algorithms, we obtain lower bounds for the performance of our algorithms. Finally, we implement our algorithms and run simulations to gather numerical results and compare our algorithms and theoretical approximation results under different sets of system parameters.
Various conceptual and descriptive models of conversational search have been proposed in the literature -- while useful, they do not provide insights into how interaction between the agent and user would change in response to the costs and benefits of the different interactions. In this paper, we develop two economic models of conversational search based on patterns previously observed during conversational search sessions, which we refer to as: Feedback First where the agent asks clarifying questions then presents results, and Feedback After where the agent presents results, and then asks follow up questions. Our models show that the amount of feedback given/requested depends on its efficiency at improving the initial or subsequent query and the relative cost of providing said feedback. This theoretical framework for conversational search provides a number of insights that can be used to guide and inform the development of conversational search agents. However, empirical work is needed to estimate the parameters in order to make predictions specific to a given conversational search setting.
The individual data collected throughout patient follow-up constitute crucial information for assessing the risk of a clinical event, and eventually for adapting a therapeutic strategy. Joint models and landmark models have been proposed to compute individual dynamic predictions from repeated measures to one or two markers. However, they hardly extend to the case where the complete patient history includes much more repeated markers possibly. Our objective was thus to propose a solution for the dynamic prediction of a health event that may exploit repeated measures of a possibly large number of markers. We combined a landmark approach extended to endogenous markers history with machine learning methods adapted to survival data. Each marker trajectory is modeled using the information collected up to landmark time, and summary variables that best capture the individual trajectories are derived. These summaries and additional covariates are then included in different prediction methods. To handle a possibly large dimensional history, we rely on machine learning methods adapted to survival data, namely regularized regressions and random survival forests, to predict the event from the landmark time, and we show how they can be combined into a superlearner. Then, the performances are evaluated by cross-validation using estimators of Brier Score and the area under the Receiver Operating Characteristic curve adapted to censored data. We demonstrate in a simulation study the benefits of machine learning survival methods over standard survival models, especially in the case of numerous and/or nonlinear relationships between the predictors and the event. We then applied the methodology in two prediction contexts: a clinical context with the prediction of death for patients with primary biliary cholangitis, and a public health context with the prediction of death in the general elderly population at different ages. Our methodology, implemented in R, enables the prediction of an event using the entire longitudinal patient history, even when the number of repeated markers is large. Although introduced with mixed models for the repeated markers and methods for a single right censored time-to-event, our method can be used with any other appropriate modeling technique for the markers and can be easily extended to competing risks setting.
Markov Chain Monte Carlo (MCMC) is one of the most powerful methods to sample from a given probability distribution, of which the Metropolis Adjusted Langevin Algorithm (MALA) is a variant wherein the gradient of the distribution is used towards faster convergence. However, being set up in the Euclidean framework, MALA might perform poorly in higher dimensional problems or in those involving anisotropic densities as the underlying non-Euclidean aspects of the geometry of the sample space remain unaccounted for. We make use of concepts from differential geometry and stochastic calculus on Riemannian manifolds to geometrically adapt a stochastic differential equation with a non-trivial drift term. This adaptation is also referred to as a stochastic development. We apply this method specifically to the Langevin diffusion equation and arrive at a geometrically adapted Langevin dynamics. This new approach far outperforms MALA, certain manifold variants of MALA, and other approaches such as Hamiltonian Monte Carlo (HMC), its adaptive variant the no-U-turn sampler (NUTS) implemented in Stan, especially as the dimension of the problem increases where often GALA is actually the only successful method. This is evidenced through several numerical examples that include parameter estimation of a broad class of probability distributions and a logistic regression problem.
The COVID-19 pandemic continues to have a devastating effect on the health and well-being of the global population. A critical step in the fight against COVID-19 is effective screening of infected patients, with one of the key screening approaches being radiological imaging using chest radiography. Motivated by this, a number of artificial intelligence (AI) systems based on deep learning have been proposed and results have been shown to be quite promising in terms of accuracy in detecting patients infected with COVID-19 using chest radiography images. However, to the best of the authors' knowledge, these developed AI systems have been closed source and unavailable to the research community for deeper understanding and extension, and unavailable for public access and use. Therefore, in this study we introduce COVID-Net, a deep convolutional neural network design tailored for the detection of COVID-19 cases from chest radiography images that is open source and available to the general public. We also describe the chest radiography dataset leveraged to train COVID-Net, which we will refer to as COVIDx and is comprised of 5941 posteroanterior chest radiography images across 2839 patient cases from two open access data repositories. Furthermore, we investigate how COVID-Net makes predictions using an explainability method in an attempt to gain deeper insights into critical factors associated with COVID cases, which can aid clinicians in improved screening. By no means a production-ready solution, the hope is that the open access COVID-Net, along with the description on constructing the open source COVIDx dataset, will be leveraged and build upon by both researchers and citizen data scientists alike to accelerate the development of highly accurate yet practical deep learning solutions for detecting COVID-19 cases and accelerate treatment of those who need it the most.
We consider the task of learning the parameters of a {\em single} component of a mixture model, for the case when we are given {\em side information} about that component, we call this the "search problem" in mixture models. We would like to solve this with computational and sample complexity lower than solving the overall original problem, where one learns parameters of all components. Our main contributions are the development of a simple but general model for the notion of side information, and a corresponding simple matrix-based algorithm for solving the search problem in this general setting. We then specialize this model and algorithm to four common scenarios: Gaussian mixture models, LDA topic models, subspace clustering, and mixed linear regression. For each one of these we show that if (and only if) the side information is informative, we obtain parameter estimates with greater accuracy, and also improved computation complexity than existing moment based mixture model algorithms (e.g. tensor methods). We also illustrate several natural ways one can obtain such side information, for specific problem instances. Our experiments on real data sets (NY Times, Yelp, BSDS500) further demonstrate the practicality of our algorithms showing significant improvement in runtime and accuracy.